Our lab has published a new preprint! It has been observed that in the state of transcriptional activation, distinctive higher-order genomic structures and protein aggregates are formed, furthermore, the viscosity around genes increases, and the interaction time between enhancers and promoters is prolonged. This research was conducted in collaboration with Drs. Hiroaki Ohishi (in our lab), Soya Shinkai (RIKEN), Shuichi Onami (RIKEN), Kazufumi Hosoda (Ansanga Labs), and Yasuyuki Ohkawa (MIB, Kyushu U.)!
https://www.biorxiv.org/content/10.1101/2023.11.27.568629v1
Recent imaging analyses have revealed that transcription is a dynamic process that switches between an active state, where genes are continuously transcribed by RNA Polymerase II, and an inactive state where transcription does not occur. This is known as transcriptional bursting and is a universal phenomenon observed across many species and cell types. It has been shown that this transition involves the interaction between enhancers and promoters, as well as the assembly of transcriptional regulatory factors.
転写因子は、遺伝子のプロモーター領域や、転写を調節するエンハンサーに結合します。これにより、RNAポリTranscription factors bind to the promoter regions of genes and enhancers that regulate transcription. This recruitment of transcription-related factors such as RNA Polymerase II and BRD4. These factors often contain many Intrinsically Disordered Regions (IDRs) and tend to form condensates. This leads to the initiation of continuous transcription by RNA Polymerase. However, the extent to which the interactions between enhancers and promoter regions are stable has not yet been fully elucidated.

On the other hand, analyses using live imaging, single-cell Hi-C techniques, and DNA-FISH have shown that higher-order genomic structures, including the interactions between enhancers and promoters, are not stable but rather dynamically changing. For example, even within the same cell type, the position of chromosomes in individual cells is not constant, and the higher-order genomic structure is dynamic, with recognized diversity in structure between cells. How this dynamic nature of higher-order genomic structures contributes to transcriptional bursting is not yet clear.

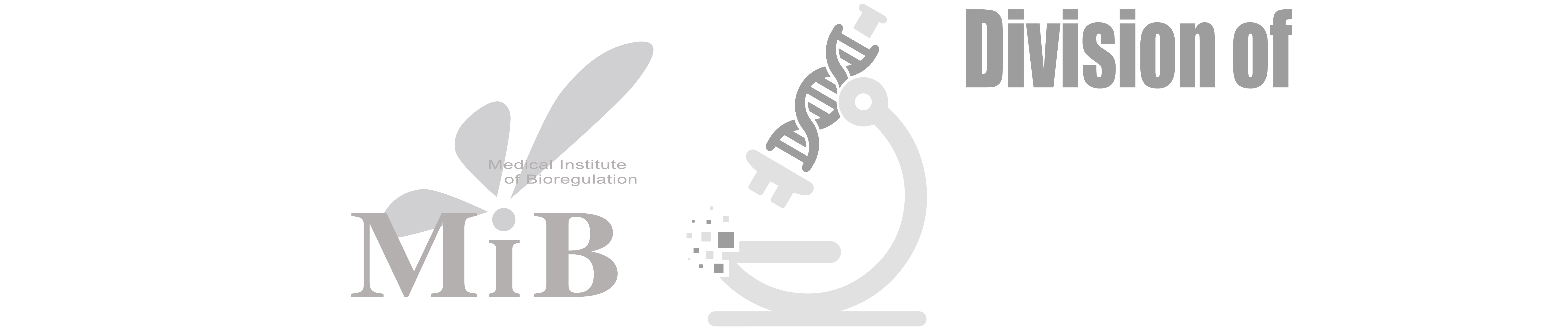
To answer this question, we adopted the RNA/DNA/IF-seqFISH technology developed by Long Cai and his team at the California Institute of Technology. Using this technique, we sequentially visualized the localization of 80 types of RNA molecules, 120 genomic regions, and 20 types of proteins along with their post-translational modifications in mouse ES cells.
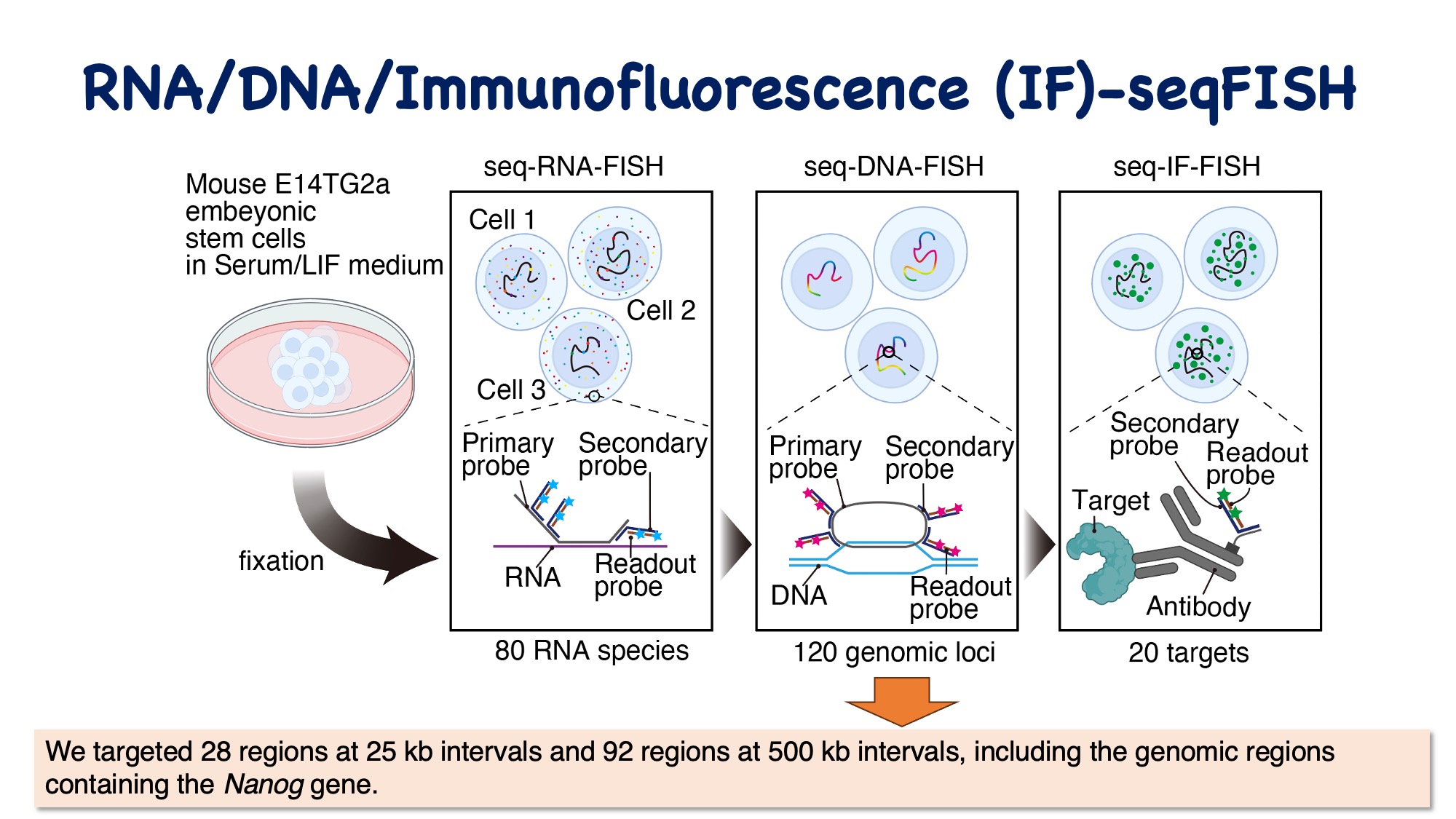
Comparing the higher-order genomic structures when Nanog is in an active and inactive state revealed structural differences. Notably, in the transcriptionally active state, a distal enhancer located approximately 190 kb upstream of Nanog was found to be positioned near the Nanog gene. Furthermore, according to IF-seqFISH data, it was observed that transcription-related factor condensates are formed around the genes in the active state. These factors are suggested to influence the interaction between enhancers and promoters.
To this end, we utilized the PHi-C technology developed by Dr. Soya Shinkai (RIKEN), and based on the data obtained from DNA-seqFISH, we estimated the dynamics of the higher-order genomic structure in the active/inactive states of Nanog. The results revealed that in the active state, there is a higher viscosity around genes compared to the inactive state. Furthermore, this suggests that in the active state, the interaction time between enhancers and promoters is more than doubled compared to the inactive state, indicating more efficient information transfer between enhancers and promoters.